Variability in IGH antibody genes influences the response to SARS-CoV-2
The COVID-19 pandemic has highlighted the importance of research focused on our ability to control infections and to explore why certain individuals respond differently than others to disease and vaccination. Recent COVID-19 research has contributed valuable information about how immune-related genes influence the ability to control SARS-CoV-2 infection. While some genes are connected with increased risk of serious infection, e.g. inborn deficiencies in type I interferon, other genes may confer protective effects against the development of severe COVID-19 disease, e.g. archaic-derived isoform of oligoadenylate synthetase (OAS), which is an interferon-induced effector molecule. These studies focused on explaining how variations in our innate immune system influence our control of SARS-CoV-2. So far, much less is known about how genetic variations underpinning our adaptive immune responses influence how we react to viral infections and our ability to form immunological memory to viruses such as SARS-CoV-2.
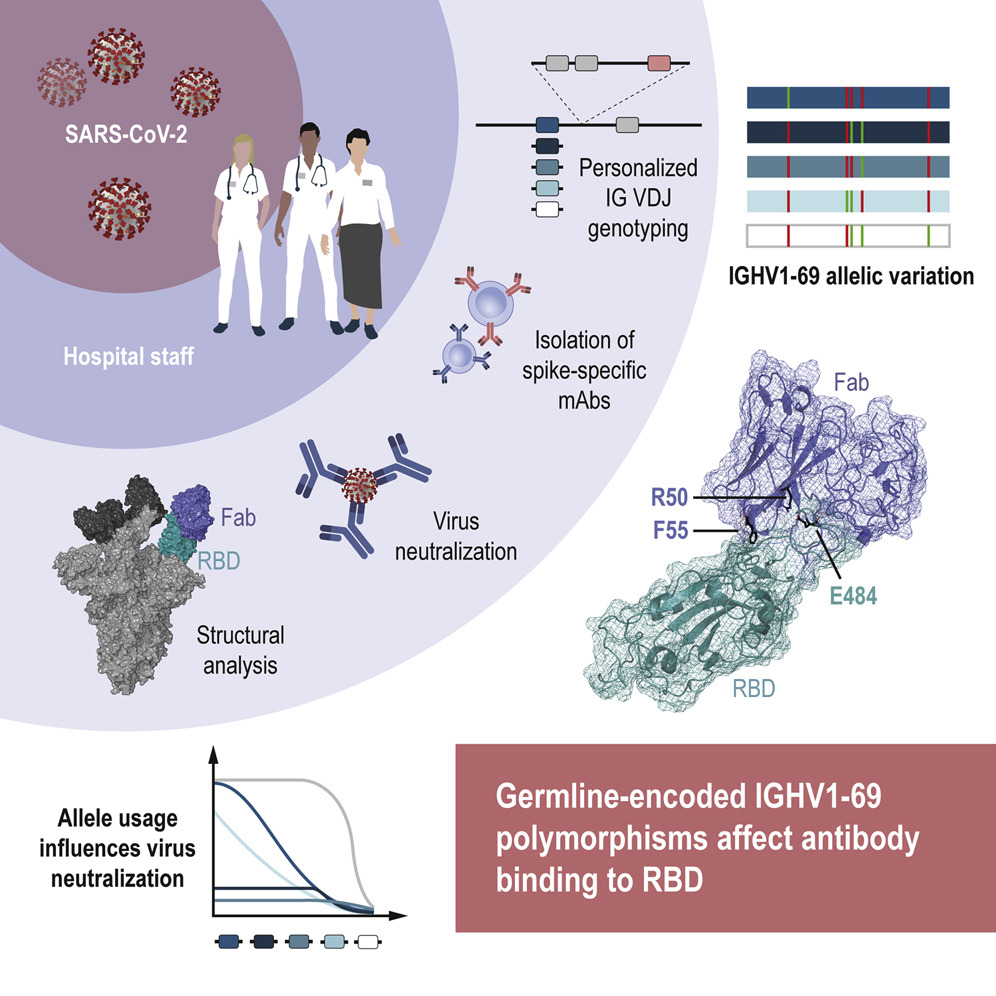
New studies show that immunoglobulin heavy chain (IGH) genes, specifically IGHV1-69, is used by many S-specific SARS-CoV-2 neutralizing antibodies. The IGHV1-69 gene is known for being both highly polymorphic and subject to copy number variation, features that shape different person’s naïve B cell repertoires. As such, it is likely that this variation has implications for the human response to infection, and vaccination.
In a recently published article in Immunity, researchers from Karolinska Institutet (First author: Pradeepa Pushparaj, Corresponding author: Gunilla B Karlsson Hedestam) studied the role of IGH germline gene variation in antibody response against SARS-CoV-2. They isolated SARS-CoV-2 spike-specific monoclonal antibodies from a previously collected cohort of convalescent healthcare workers (N=14, seropositive for S-IgG) and selected a set of potently neutralizing IGHV1-69-using antibodies. They used the IG genotyping tool IgDiscover (learn more about the software here) and inferred haplotype analysis to study the variation in IGHV allele composition between the study participants. While one participant was homozygous for IGHV1-69∗01, nine had two or three different alleles of this gene and two were found to have four different IGHV1-69 alleles. Six different alleles, e.g. IGHV1-69∗20, were found among the participants.
The team focused on an individual who had three IGHV1-69 alleles from whom they cloned the IGHV1-69∗20-using CAB-I47 antibody, and two highly similar antibodies that were isolated from an independent donor. They then engineered the three IGHV1-69∗20-using mAbs to assess if germline-encoded polymorphisms that characterize the IGHV1-69*20 allele were required for the neutralization activity of the antibodies. They found this to be the case. By using cryo-EM to analyze structural data, they found that the CAB-I47 Fab, in complex with the SARS-CoV-2 spike, docked into the receptor-binding domain (RBD) of the virus, with two germline-encoded polymorphisms in IGHV1-69, R50 and F55, being required for the high affinity RBD interaction. They also confirmed this finding experimentally.
Gunilla B. Karlsson Hedestam, the corresponding author says “Antibody genetics is a notoriously complex field. Thanks to new experimental methods and our computational tool, IgDiscover, we can now resolve the fine details of antibody genes. These studies reveal a surprising level of genetic variability among humans, which influence how we respond to infections”.
In summary, this study showed that human IGH antibody genes are highly variable and germline-encoded residues specific to given IGHV1-69 alleles can shape the neutralizing antibody response to SARS-CoV-2. These results demonstrate that genetic differences can influence our antibody response to SARS-CoV-2 and thus also shape long-lived memory B cell responses induced by infection or vaccination.
Data
Adhering to Open Science and FAIR the researchers have shared sequence data in GenBanka and data in several repositories such as ProteomeXchange, PDB, EMD and SciLifeLab Data Repository.
- HC (VDJ) and LC (VJ) sequences of neutralizing mAbs available in GenBank through accession id OP497961 - OP497964 and ON086918 - ON086947.
- The repertoire sequence data is available in Figshare hosted by SciLifeLab.
- Cryo-EM structures of CAB-I47 Fabs in complex with the spike trimer are deposited Protein Data Bank (8A99, 8A94, and 8A96) and Electron Microscopy Data Bank (EMD-15273, EMD-15269, and EMD-15271).
- Mass spectrometry raw files and HDX analysis, deposited on ProteomeXchange Consortium via PRIDE. Can be accessed by PXD031945.
- The IgDiscover software can be found here (The plot allele module and the rep-seq analysis tools can be found under IgDiscover v0.12.4.dev22+g0bc3365).
- Supplemental Tables are available here.
Article
DOI: 10.1016/j.immuni.2022.12.005
Pushparaj, P., Nicoletto, A., Sheward, D. J., Das, H., Castro Dopico, X., Perez Vidakovics, L., Hanke, L., Chernyshev, M., Narang, S., Kim, S., Fischbach, J., Ekström, S., McInerney, G., Hällberg, B. M., Murrell, B., Corcoran, M., & Karlsson Hedestam, G. B. (2023). Immunoglobulin germline gene polymorphisms influence the function of SARS-CoV-2 neutralizing antibodies. In Immunity (Vol. 56, Issue 1, pp. 193-206.e7).
Funding
This project was funded the Swedish Research Council, grants from the SciLifeLab National COVID-19 Research Program financed by the Knut and Alice Wallenberg Foundation and the SciLifeLab Pandemic Laboratory Preparedness (PLP) program 2022–2023. Additional funding was provided from the Knut and Alice Wallenberg Foundation and the European Union’s Horizon 2020 research and innovation programme.
Infrastructure
Part of this work was supported by the Swedish National Infrastructure for Biological Mass Spectrometry (BioMS) and the SciLifeLab (Sweden) Integrated Structural Biology platform. All cryo-EM data were collected at the Karolinska Institutet’s 3D-EM facility, and surface plasmon resonance analysis was performed at the Protein Science Facility at Karolinska Institutet.