METTL3 localisation during SARS-CoV-2 infection could highlight new novel antiviral strategy
During recent years, the rapid development of COVID-19 treatments and vaccines have proven crucial in controlling the spread of the SARS-COV-2 virus and mitigating the effects of the COVID-19 pandemic. The COVID-19 pandemic is now considered to be at an end, with the virus becoming endemic. However, it is still important that we continue to deepen our understanding of SARS-CoV-2 pathogenesis (e.g. the basic mechanisms of how the virus infects hosts, host-virus interactions, and host response). This is not only because SARS-CoV-2 continues to circulate in populations, but also to protect against future pandemics by this or similar viruses.
The N6-Methyladenosine modification (m6A) is one of the most common cellular RNA modifications, and known to play a crucial role in the regulation of RNA metabolism during the stress response. However, little is known about how m6a expression is altered during SARS-CoV-2 infection or the impact that any changes might have. This knowledge may hold clues for novel or improved antiviral therapeutics for SARS-CoV-2. It may also increase our general knowledge about the mechanism of coronaviruses, and other potential pandemic threats.
In a recent publication in Genome Research, researchers from Sweden, in collaboration with researchers from South Korea, India, and France (First authors: Roshan Vaid and Akram Mendez, Corresponding author: Tanmoy Mondal), studied how the gene expression profile of m6A mRNA is affected during and after COVID-19 infection.
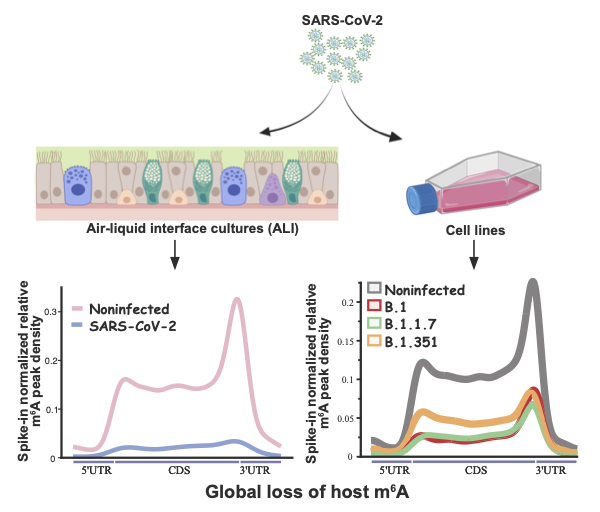
Vaid and Mendez et al used Vero cells and air/liquid interface (ALI) cultures of human airway epithelia to study SARS-CoV-2 infection. The researchers chose to study three SARS-CoV-2 variants; B.1, B.1.1.7, and B.1.351. All three SARS-CoV-2 variants showed changes in the expression of genes related to RNA catabolism, including m6A readers and erasers. Whilst m6A was found to be abundantly detected in viral RNA, infection with the SARS-CoV-2 variants studied caused a loss of m6A in cellular RNAs.
The methyltransferase METTL3/METTL14 complex, is the key cellular enzyme complex responsible for depositing m6A modifications. It is normally localised in the nucleus. The initial finding of the study by Vaid and Mendez et al, i.e. that there is a global loss of m6A peaks during SARS-CoV-2 infection, could indicate that the function of the METTL3/METTL14 is altered during SARS-CoV-2 infection. Vaid and Mendez et al. continued to investigate this finding and found that, during SARS-CoV-2 infection, METTL3 is partially relocalised from the nucleus to the cytoplasm. This change in localisation from nucleus to was more prominent during infection with the SARS-CoV-2 B.1 and B.1.1.7 variants, than with the SARS-CoV-2 B.1.351 variant. These results were consistent with the finding that there was a greater reduction in the level of m6a during infection with SARS-CoV-2 B.1 and B.1.1.7, compared to infection with B.1.351. In addition, the researchers were able to restore the localisation of the METTL3 by inhibiting the export protein XPO1. This inhibition also caused a recovery of m6A in cellular RNA, and increased mRNA expression.
“We were surprised by the drastic loss of host cell m6A RNA modification during SARS-CoV-2 infection. We do not know how long it takes to regain m6A post-infection. SARS-COV-2 may leave a long-lasting mark in the infected cells and this might provide clues to why some people have chronic symptoms that persist long after COVID.” says corresponding author Tanmoy Mondal.
In conclusion, future pandemic preparedness efforts will require the development of new antiviral strategies and antiviral drugs. The findings from Vaid and Mendez et al highlight how SARS-CoV-2 perturbs the m6A RNA modification pathway to deregulate cellular RNAs, and thereby limits stress granule formation. METTL3 localisation during SARS-CoV-2 infection could highlight a new novel antiviral strategy for treatment of COVID-19. Further studies into this potential novel strategy are warranted.
Data
The researchers first published their articles as a preprint on 22nd December 2022 on BioRXiv. They also adhere to Open Science by sharing data and code openly.
- All raw and processed sequencing data generated in this study have been submitted to the NCBI Gene Expression Omnibus (GEO) under accession number GSE188477.
- The source code used for the analysis of data in this study is available on GitHub and in supplementary data.
Article
Vaid, R., Mendez, A., Thombare, K., Burgos-Panadero, R., Robinot, R., Fonseca, B. F., Gandasi, N. R., Ringlander, J., Hassan Baig, M., Dong, J.-J., Cho, J. Y., Reinius, B., Chakrabarti, L. A., Nystrom, K., & Mondal, T. (2023). Global loss of cellular m6A RNA methylation following infection with different SARS-CoV-2 variants. In Genome Research (Vol. 33, Issue 3, pp. 299–313).
Funding
This work was funded by grants from the Swedish Research Council, Sweden-South Korea COVID-19 grant from the Swedish Research Council, Svenska Läkaresällskapet; Kungl. Vetenskaps- och Vitterhets-Samhället (KVVS), as well as several international funders.
Infrastructure
The core facility at Novum, BEA, Bioinformatics and Expression Analysis has supported with sequencing, and the Proteomic core facility at Gothenburg University supported with LC-MS/MS. Swedish National Infrastructure for Computing (SNIC) at UPPMAX has supported computations and data handling.