New data highlight exploring a new analysis pipeline for mRNA-Seq and ATAC-Seq data
August 19, 2024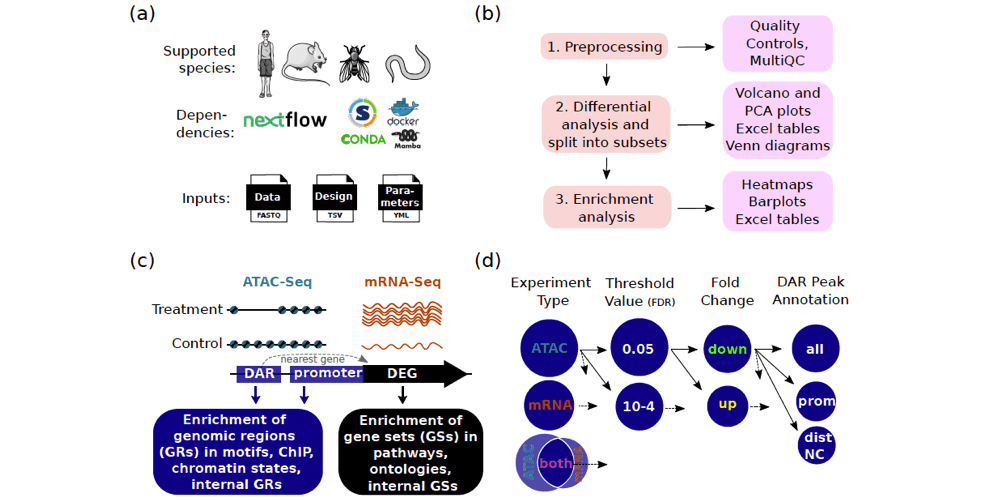
The latest data highlight is entitled ‘New pipeline for mRNA-Seq and ATAC-Seq analysis allows for biological insights without in-depth bioinformatics skills’, and is based on recent research by Salignon et al. (2024).
Both mRNA-Seq (which measures gene expression), and ATAC-Seq (which measures chromatin accessibility) are frequently used in omics research. Multiple pipelines are available to analyse either mRNA-Seq or ATAC-Seq data. However, they typically have limited options for downstream analyses, such as enrichment analysis and integration with mRNA-Seq data. The Cactus (Chromatin accessibility and transcriptomics unifying software) pipeline, developed by Salignon et al. (2024) has multiple unique functionalities compared to other ATAC-Seq data analysis tools. This includes, for example, splitting the differential analyses results into subsets, enrichment in chromatin states and ChIP-Seq binding sites, and the creation of customised heatmaps and pdf reports. Unlike other pipelines, the Cactus pipeline therefore enables researchers to perform efficient and reproducible analyses of interrelated changes in chromatin accessibility and gene expression, without the need for advanced bioinformatics skills. The Cactus pipeline is available for use on multiple NAISS resources, including Rackham, Bianca, Miarka, and Snowy.
For more details, see the data highlight: ‘New pipeline for mRNA-Seq and ATAC-Seq analysis allows for biological insights without in-depth bioinformatics skills’. Other relevant articles that might be of interest are also shown at the bottom of the data highlight.
To get your work featured as a data highlight, please get in touch with us to suggest it.